department of molecular biosciences, evanston, il 60208
| |
| home |
| research |
| people |
| gallery |
| knowledgebase |
| affiliations |
| Research |
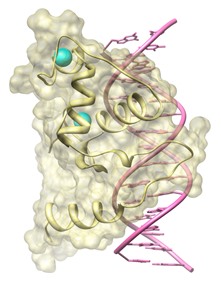
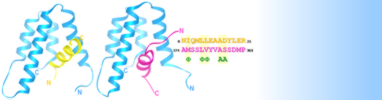
Research in the Radhakrishnan lab focuses on the molecular mechanisms of eukaryotic transcription regulation with emphasis on how transcription factors engage with DNA, recruit specific coactivators and corepressors via intrinsically disordered transactivation or transrepression domains and how multi-protein coactivator/corepressor-bearing chromatin-modifying complexes assemble and engage with chromatin. We are asking these questions in the context of (i) nuclear receptors that use atypical mechanisms to effect transcriptional activation and (ii) a cohort of related, yet functionally distinct, histone deacetylase (HDAC)-associated chromatin-modifying complexes that fundamentally impact on cellular physiology in eukaryotes. We address these questions using molecular biological, biochemical, and biophysical approaches including solution NMR spectroscopy, electron paramagnetic resonance (EPR), macromolecular X-ray crystallography, and cryogenic electron microscopy (cryoEM) as well as computational approaches including informatics and molecular dynamics (MD) simulations. Both projects have immense biological and biomedical significance as inhibitors of HDACs and nuclear receptors are targets for treating a variety of human diseases including cancer. Current projects in the lab focus on the NR4A family of nuclear receptors comprising Nur77, Nurr1, and NOR1. Each of these receptors plays important roles in metabolism, inflammation, and the proper development of dopaminergic neurons, among others. We are asking how these rather atypical receptors activate transcription via their ligand-binding domain as well as their activation domain at the N-terminus of these proteins. We are also asking whether these receptors function in a ligand-dependent or ligand-independent manner. Separately, we are asking how the evolutionarily-conserved, histone deacetylase (HDAC)-containing Sin3L/Rpd3L complex is assembled, what the precise molecular role(s) of the conserved subunits, which harbor domains of poorly characterized structure and function, are, including whether they regulate HDAC activity and how the complex engages chromatin and DNA-bound factors. Finally, we are developing the next iteration of a popular web application called MONSTER that can mine experimentally-determined structures for stabilizing interactions in macromolecular complexes. New enhancements include a JavaScript-based user interface, a database for storing and mining results, a stability predictor for mutants, and an automated tool for generating evolutionary conservation profiles. |
SELECTED PUBLICATIONS Daffern, N. and Radhakrishnan, I. (2022). A novel mechanism of coactivator recruitment by the nuclear receptor Nurr1. J. Mol. Biol., 434, 167718. Marcum, R.D., Hsieh, J., Giljen, M., Justice, E., Daffern, N., Zhang, Y. and Radhakrishnan, I. (2022). A capped Tudor domain within a core subunit of the Sin3L/Rpd3L histone deacetylase complex binds to nucleic acid G-quadruplexes. J. Biol. Chem., 298, 101558. Marcum, R.D. and Radhakrishnan, I. (2020). The neuronal transcription factor Myt1L interacts via a conserved motif with the PAH1 domain of Sin3 to recruit the Sin3L/Rpd3L histone deacetylase complex. FEBS Lett., 594, 2322-2330. Marcum, R.D. and Radhakrishnan, I. (2019). Inositol phosphates and core subunits of the Sin3L/Rpd3L histone deacetylase (HDAC) complex up-regulate deacetylase activity. J. Biol. Chem. 294, 13928-13938. Daffern, N., Chen, Z., Zhang, Y., Pick, L. and Radhakrishnan, I. (2018). Solution NMR studies of the ligand-binding domain of an orphan nuclear receptor reveal a dynamic helix in the ligand-binding pocket. Biochemistry 57, 1977-1986. Clark, M.D., Zhang, Y. and Radhakrishnan, I. (2015). Solution NMR Studies of an alternative mode of Sin3 engagement by the Sds3 subunit in the histone deacetylase-associated Sin3L/Rpd3L corepressor complex. J. Mol. Biol. 427, 3817-3823. Clark, M.D., Kumar, G.S., Marcum, R., Luo, Q., Zhang, Y., and Radhakrishnan, I. (2015). Molecular basis for the mechanism of constitutive CBP/p300 coactivator recruitment by CRTC1-MAML2 and its implications in cAMP signaling. Biochemistry 54, 5439-5446. Clark, M.D., Marcum, R., Graveline, R., Chan, C.W., Xie, R. Chen, Z., Ding, Y., Zhang, Y., Mondragón, A., David, G., and Radhakrishnan, I. (2015). Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex. Proc. Natl. Acad. Sci. USA 112, E3669-E3678. Xie, T., Zmyslowski, M., Zhang, Y., and Radhakrishnan, I. (2015). Multi-specificity of MRG domains. Structure 23, 1049-1057. Luo, Q., Viste, K., Zaa, J.C., Kumar, G.S., Tsai, W.W., Talai, A., Mayo, K.E., Montminy, M., and Radhakrishnan, I. (2012). Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc. Natl. Acad. Sci. USA 109, 20865-20870. Kumar, G.S., Chang, W., Xie, T., Patel, A., Zhang, Y., Wang, G.G., David, G., and Radhakrishnan, I. (2012). Sequence requirements for combinatorial recognition of histone H3 by the MRG15 and Pf1 subunits of the Rpd3S/Sin3S corepressor complex. J. Mol. Biol. 422, 519-531. Xie, T., Graveline, R., Kumar, G.S., Zhang, Y., Krishnan, A., David, G. and Radhakrishnan, I. (2012). Structural basis for molecular interactions involving MRG domains: Implications in chromatin biology. Structure 20, 151-160. Xie, T., He, Y., Korkeamaki, H., Zhang, Y., Imhoff, R., Lohi, O., and Radhakrishnan, I. (2011). Structure of the 30 kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding. J. Biol. Chem. 286, 27814-27824. Kumar, G.S., Xie, T., Zhang, Y., and Radhakrishnan, I. (2011). Solution structure of the mSin3A PAH2-Pf1 SID1 Complex: a Mad1/Mxd1-like interaction disrupted by MRG15 in the mammalian Rpd3S/Sin3S complex. J. Mol. Biol. 408, 987-1000. He, Y., Imhoff, R., Sahu, A., and Radhakrishnan, I. (2009). Solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex and its potential role in nucleic acid recognition. Nucleic Acids Res. 37, 2142-2152. Sahu, S.C., Swanson, K.A., Kang, R.S., Huang, K., Brubaker, K., Ratcliff, K., & Radhakrishnan, I. (2008). Conserved themes in target recognition by the PAH1 and PAH2 domains of the Sin3 transcriptional corepressor. J. Mol. Biol. 375, 1444-1456. Little, T.H., Zhang, Y., Matulis, C.K., Weck, J., Zhang, Z., Ramachandran, A., Mayo, K.E. and Radhakrishnan, I. (2005). Sequence-specific DNA recognition by Steroidogenic Factor 1: A helix at the carboxy-terminus of the DNA binding domain is necessary for complex stability. Mol. Endocrinol. 20, 831-843. Swanson, K.A., Knoepfler, P.S., Huang, K., Kang, R.S., Cowley, S.M., Laherty, C.D., Eisenman, R.N., and Radhakrishnan, I. (2004). HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 11, 738-746. Swanson, K.A., Kang, R.S., Stamenova, S.D., Hicke, L., and Radhakrishnan, I. (2003). Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 22, 4597-4606. Kang, R.S., Daniels, C.M., Francis, S.A., Shih, S.C., Salerno, W.J., Hicke, L., and Radhakrishnan, I. (2003). Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 113, 621-630. Brubaker, K., Cowley, S.M., Huang, K., Loo, L., Yochum, G.S., Ayer, D.E., Eisenman, R.N., and Radhakrishnan, I. (2000). Solution structure of the interacting domains of the Mad-Sin3 complex: Implications for recruitment of a chromatin-modifying repression complex. Cell 103, 655-665. Radhakrishnan, I., Pérez-Alvarado, G.C., Parker, D., Dyson, H.J., Montminy, M.R., and Wright, P.E. (1997). Solution structure of the KIX binding domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell 91, 741-752. |